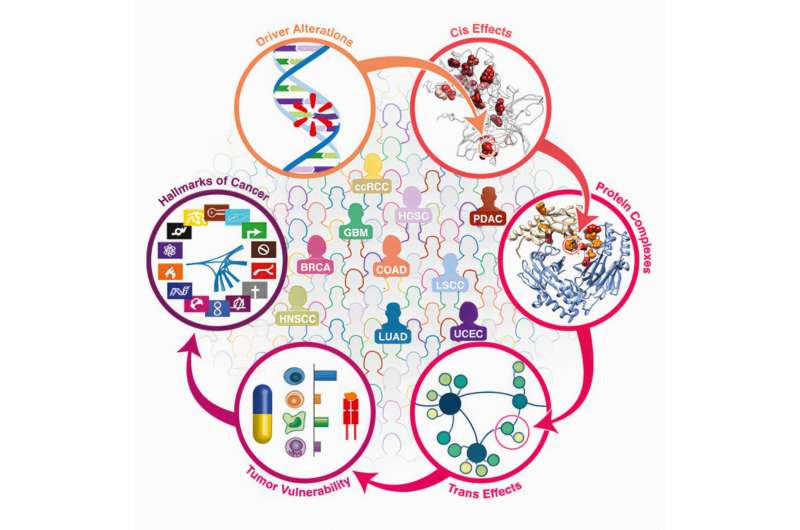
Scientists have completed a deep analysis of the proteins driving cancer across multiple tumor types, information that can’t be assessed by genome sequencing alone. Understanding how proteins operate in cancer cells raises the prospect of new therapies that block key proteins that drive cancer growth, or therapies that trigger immune responses to abnormal proteins created by cancer cells.
Led by Washington University School of Medicine in St. Louis, the Broad Institute of MIT and Harvard, Brigham Young University and other institutions around the world, the Clinical Proteomic Tumor Analysis Consortium investigates key proteins driving cancer and how they’re regulated.
The findings were published Aug. 14 in a set of papers in the journals Cell and Cancer Cell.
“In our efforts to develop better cancer therapies, this new analysis of the proteins driving tumor growth is the next step after cancer genome sequencing,” said senior author Li Ding, Ph.D., the David English Smith Distinguished Professor of Medicine at Washington University.
“Through our past work sequencing the genomes of cancer cells, we identified almost 300 genes driving cancer. Now, we are studying the details of the machinery these cancer genes set in motion—the proteins and their regulatory networks that actually do the work of causing uncontrolled cell division. We are hopeful this analysis will serve as an important resource for cancer researchers seeking to develop new treatments for many tumor types.”
The researchers analyzed about 10,000 proteins involved in 10 different types of cancer. Ding emphasized the importance of the sheer volume of data in this type of analysis; many of these important cancer-driving proteins are rare in any single cancer and could not have been identified had the tumor types been studied individually. The analysis included two different types of lung cancer as well as colorectal, ovarian, kidney, head and neck, uterine, pancreatic, breast and brain cancers.
“Many of these proteins driving cancer are found in multiple tumor types but at low frequency,” said Ding, also a research member of Siteman Cancer Center at Barnes-Jewish Hospital and Washington University School of Medicine. “When we analyze many cancer types together, we increase the power we have to detect important proteins that are causing the cancer to grow and spread. A combined analysis also allows us to pinpoint the major common mechanisms driving cancers across types.”
Beyond the function of individual proteins, such data also allow the researchers to understand how proteins interact with one another to fuel cancer growth. If the levels of two proteins correlate with one another—for example, when one is present at high levels and the other always is as well—this can indicate that the two proteins act as partners. Disrupting the interaction may be a promising way to block tumor growth.
The studies, including one co-led by Ding and Gad Getz, Ph.D., of the Broad Institute, also revealed different ways proteins can be chemically altered to change their function. The researchers documented how such chemical changes—processes called acetylation and phosphorylation—can alter DNA repair, change immune responses, and modify how DNA is folded and packaged, among other important molecular changes that can play roles in driving cancer.
The research also shed light on the effectiveness of immunotherapies. Immunotherapies such as checkpoint inhibitors often work best in cancers with a lot of mutations, but even then, they don’t work for all patients. The researchers found that high numbers of mutations don’t always result in an abundance of abnormal proteins, which is what the immune system targets when attacking a tumor.
“For some cancers, even with mutations having the potential to generate tumor antigens, if there is no abnormal protein expressed, or very little, such mutations may not be targetable for treatment,” Ding said. “This could be an explanation for why some patients don’t respond to immunotherapy, even when it seems like they should. As such, our proteomics investigation covering the expression profiles of tumor antigens are particularly useful for designing new immunotherapy targeting selected mutations.”
In another study, Ding’s team identified patterns of DNA methylation, another chemical alteration that can influence the way genes are expressed. Such patterns can be key cancer drivers. In one important finding, the team identified a molecular switch that suppresses the immune system in certain tumor types.
The final paper in the set of four studies makes the data and analysis resources used by the consortium available to the wider research community.
“In general, this thorough proteomic and chemical modification analysis across many cancer types—combined with our longstanding knowledge of cancer genomics—provides another layer of information that we hope can help answer many ongoing questions about how cancer grows and manages to evade many of our best treatments,” she said.
More information:
Yize Li et al, Pan-cancer proteogenomics connects oncogenic drivers to functional states, Cell (2023). DOI: 10.1016/j.cell.2023.07.014
Yifat Geffen et al, Pan-cancer analysis of post-translational modifications reveals shared patterns of protein regulation, Cell (2023). DOI: 10.1016/j.cell.2023.07.013
Wen-Wei Liang et al, Integrative multi-omic cancer profiling reveals DNA methylation patterns associated with therapeutic vulnerability and cell-of-origin, Cancer Cell (2023). DOI: 10.1016/j.ccell.2023.07.013
Yize Li et al, Proteogenomic data and resources for pan-cancer analysis, Cancer Cell (2023). DOI: 10.1016/j.ccell.2023.06.009
Provided by
Washington University School of Medicine
Citation:
Scientists reveal how proteins drive growth of multiple cancer types (2023, August 15)
retrieved 15 August 2023
from https://phys.org/news/2023-08-scientists-reveal-proteins-growth-multiple.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.









