A research group led by Prof. Liang Haojun and researcher Bao Jianqiang from the University of Science and Technology of China (USTC) proposed a novel functional double-stranded DNA (dsDNA) donor that greatly enhances the efficiency of gene knock-in.
Their study, titled “Efficient precise integration of large DNA sequences with 3′-overhang dsDNA donors using CRISPR/Cas9,” was published in the Proceedings of the National Academy of Sciences (PNAS) on May 22, 2023.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) have revolutionized genome manipulation for research and gene therapy purposes, relying on homology-directed repair (HDR) pathways that involve exogenous donor templates with homology arms for precise gene knock-in. However, the HDR pathway’s efficiency decreases significantly when working with gene-sized DNA donors.
Current methods of enhancing gene knock-in efficiency, such as chemical modifications or covalent linking of donors to the CRISPR system, present operational difficulties and high costs. Therefore, there is an urgent need for further optimization of gene knock-in donors to facilitate the application of CRISPR/Cas9-mediated gene knock-in in life sciences and clinical treatments.
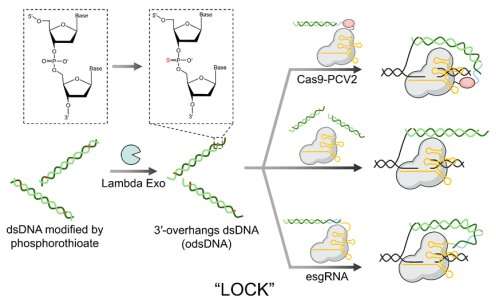
In response to this need, Prof. Liang’s group conducted extensive investigations into the mechanisms of single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA) knock-in donors. Leveraging their expertise in nucleic acid chemistry, they integrated ssDNA and dsDNA into a single donor, resulting in the design of a novel dsDNA structure with a 3′-overhang (odsDNA). This odsDNA was then used as a donor for CRISPR/Cas9-mediated large fragment gene knock-in, termed LOCK (Long dsDNA with 3′-Overhangs mediated CRISPR Knock-in).
The study introduces an economical and universal method for preparing odsDNA donors with arbitrary 3′-overhang lengths. This latest breakthrough builds upon the former achievements of Prof. Liang’s research group in gene editing. They previously enhanced gene knock-in efficiency using chemically modified 5′-ends of dsDNA donors and developed an RNA-mediated CRISPR-dCas9 transcriptional regulation system. The desired gene sequence was then obtained by using primers with phosphorothioate modifications through PCR amplification. Subsequent digestion with Lambda exonuclease produced odsDNA with a 3′-overhang.
Comparisons between gene-sized DNA donors (1.1 kb and 2.5 kb) revealed that odsDNA significantly improved targeted precise integration efficiency, achieving up to 4.3-fold enhancement compared to conventional dsDNA donors. Furthermore, odsDNA maintained lower insertion-deletion rates and off-target events across multiple genomic loci in mammalian cells. By combining 3′-single-stranded overhangs with ligation techniques, the knock-in efficiency increased 5.2-fold, compared to dsDNA donors.
The LOCK strategy demonstrates remarkable advantages in gene knock-in and holds the potential to replace ssDNA or dsDNA donors, enabling large fragment gene knock-in which was previously unachievable.
More information:
Wenjie Han et al, Efficient precise integration of large DNA sequences with 3′-overhang dsDNA donors using CRISPR/Cas9, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2221127120
Provided by
University of Science and Technology of China
Citation:
Double-stranded DNA donor with novel structure boosts gene knock-in efficiency (2023, July 13)
retrieved 16 July 2023
from https://phys.org/news/2023-07-double-stranded-dna-donor-boosts-gene.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.









